Complex Association (Merge or Split):
The aim of this part is to show how subunits or macromolecules bind one to each other.
The key point is to have a good knowledge of the protein's structure (namely the pdb file) that will be used. For this purpose, use the preview plug-in
to check out the orientation of each chain(s) and of the bound complex ("All"). The specific options for complex
animation are: the movement and priority of each chain(s) and the
camera type (simple, wide, ultra-wide, zoom, rotX, rotY).
Quick start
a) Set
3CRO in the "enter a PDB Id" field. The pdb code is related to a protein-dna complex.
b) In the Default Representation part:
- Title: DNA-Protein complex
- Display: keep all values to default
- Scene: keep all values to default
- Picture: change only the image format to gif
c) Animation
- Scenarii: change to "Complex Merge"
- Camera: change to "wide"
- Render: megapov
- Frames: 20, step 1, delay 20
d) Orientation panel: we will now define the scenario. The 3*3 panel represents the different possible positions and movements for each chain. Use the preview orientation to help you. Thus click on the Preview orientations button. A jmol applet is launch. You could see the diffent chains of 3CRO (colored by different colors). In 3CRO there is 4 chains: A,B for the DNA (lightblue and yellow) and L,R for the bounded protein (blue and green).
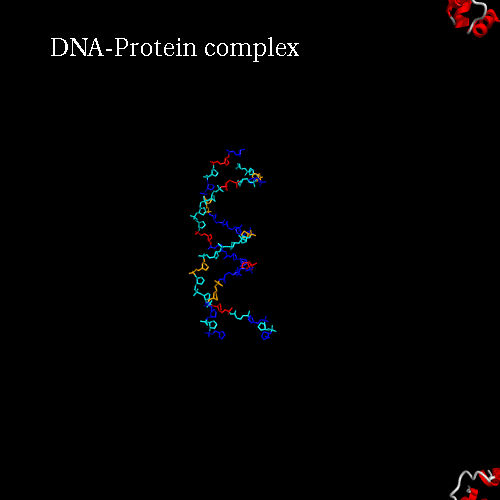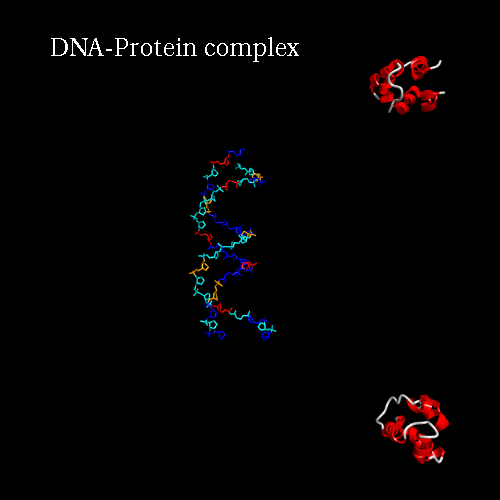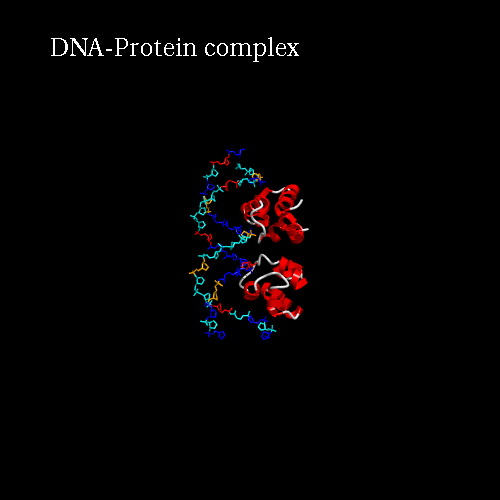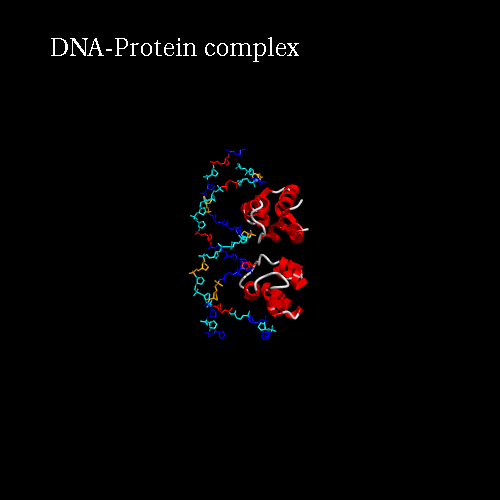
Our idea is to create a animation showing the protein while binding to the DNA (merge). In order to position the bounded form at the center of the scene, we kept the "chains" field to "All", which corresponds to the bounded form (ADN+Protein), in the central cell of the 3*3 panel. But we reoriented it to get a better view by changing the rotation to : Xrot -81, Yrot 69 and Zrot 19. We want the L and R chains to move from the right to the center of the scene, therefore we set "L" and "R" in the "chains" field of the bottom and upper right cells respectively. To simplify the animation we didn't change the orientations of
chains L and R. Once again click on the Preview orientations to find a right orientation for each partnairs.
d) Process : launch the procedure to create the animated gif, just clic the Process button.
Here is what you should get:
|
Engine : Dino / Render : Megapov
|
Engine : YASARA / Render : Megapov
|
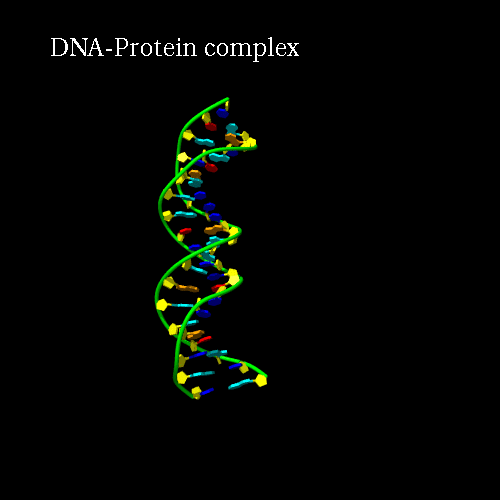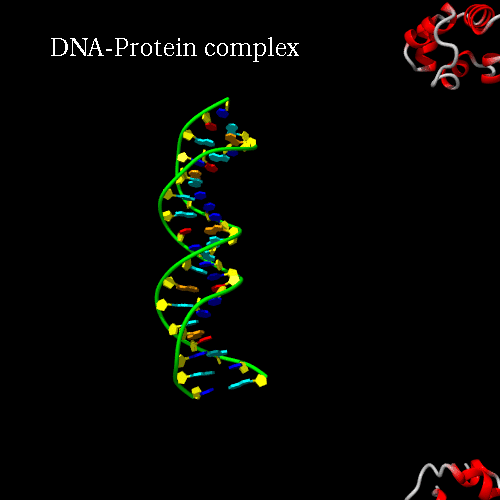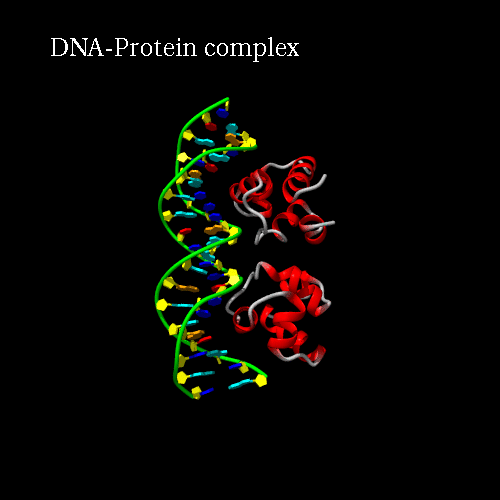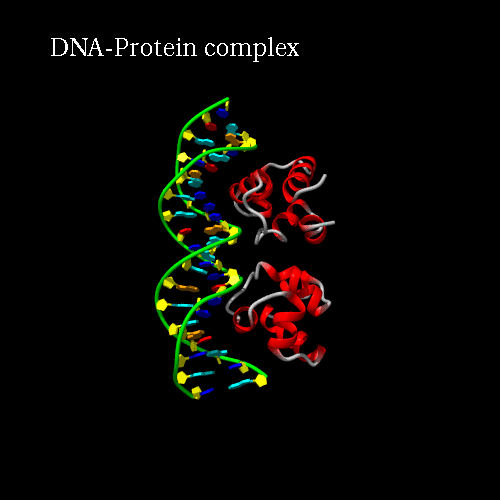
|
|
Now we will see how use the capabilities of the 3*3 panel. Previously we saw how to define the behavior of different chains by placing them in different cells. In the form, pay attention to the "Pr:" field (for Priority). The purpose of this field is to define the order of the moving chains. Namely, if we want that chain A moves first, and then chain B follows, we have to define "Pr" of chain A to "1" and "Pr" of chain B to "2".
So just follow the form below:
You probably noticed that we applied rotations to both protein subunits, just by setting angle values in the corresponding fields (Chain R Xrot 15. Yrot 55. and chain L Xrot 25. Yrot 45.)
You should get this nice animation:
Here is the same scene (representation and scenarii, without surface) but with the different camera options : simple, wide, ultra-wide and zoom (for comparison we also show the result using the YASARA engine):
Here are some other examples of animation of macromolecular complexes :
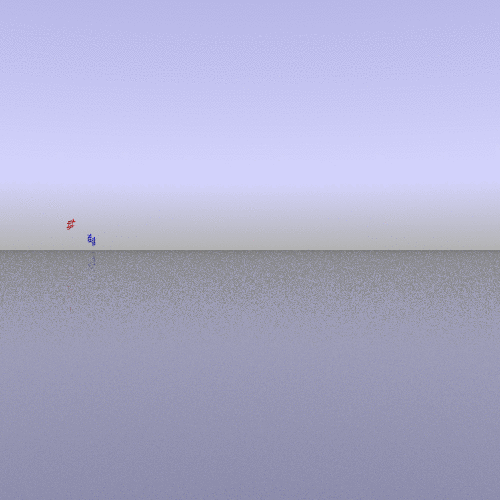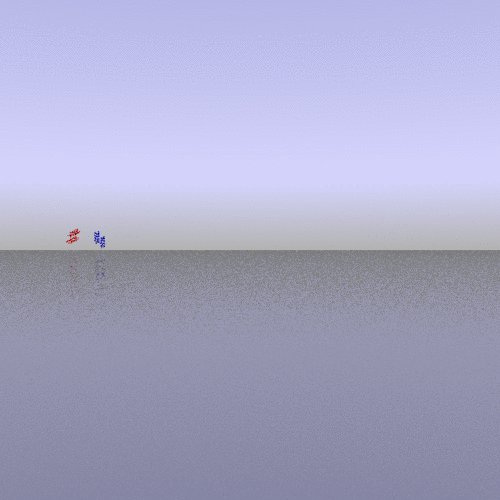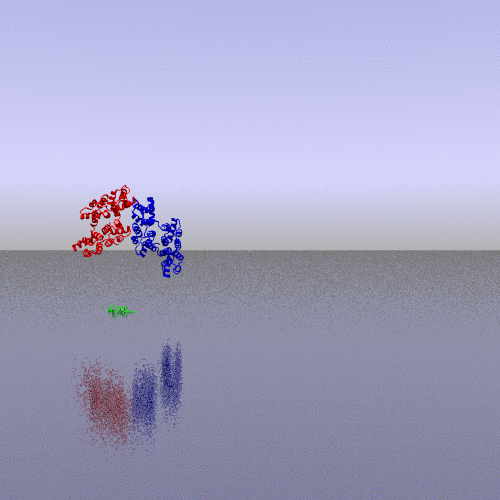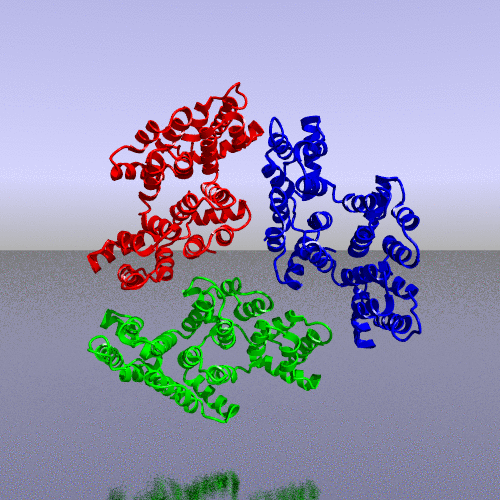
PDB code 1ANX, annexin trimer form, displayed as backbone cartoon colored according to "Chains"
with a black background, and a scenario of type "Complex Merge" with 10 frames.
Here is the 3*3 panel:
| chain B Pr:2
| None
| chain A Pr:1
|
| None
| All
| None
|
| None
| chain C Pr:3
| None
|
|
Engine : Dino / Render : Megapov
|
Engine : YASARA / Render : Megapov
|
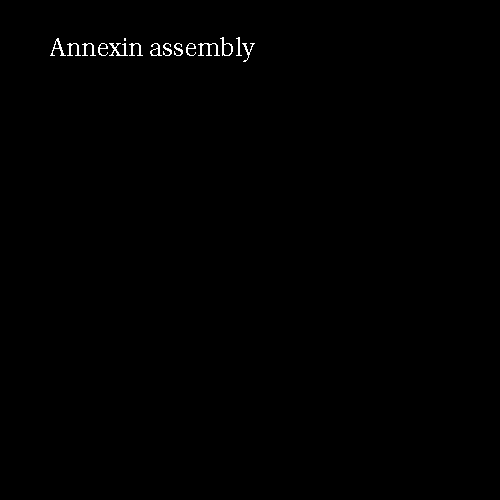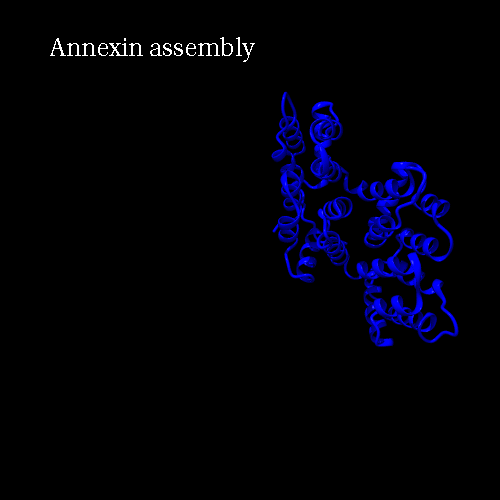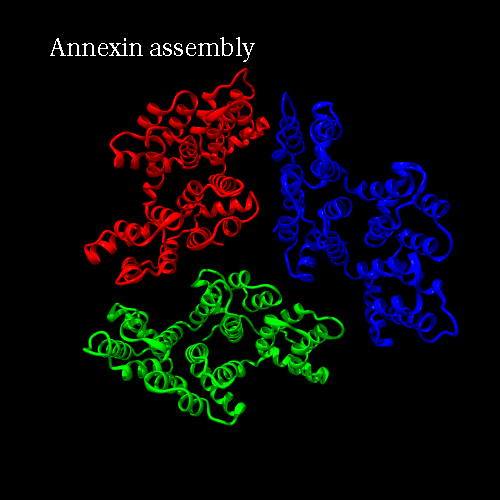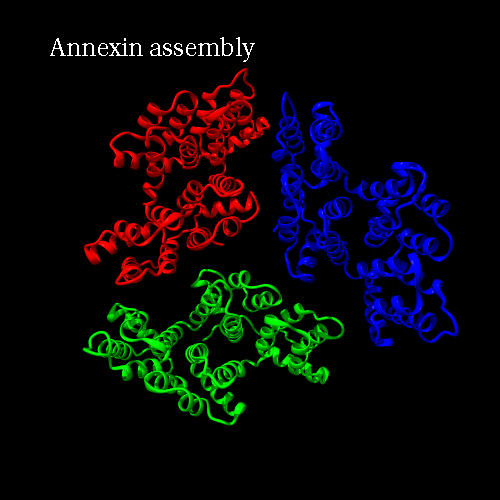
|
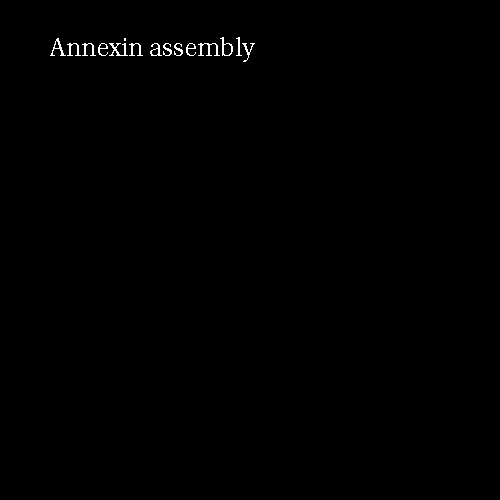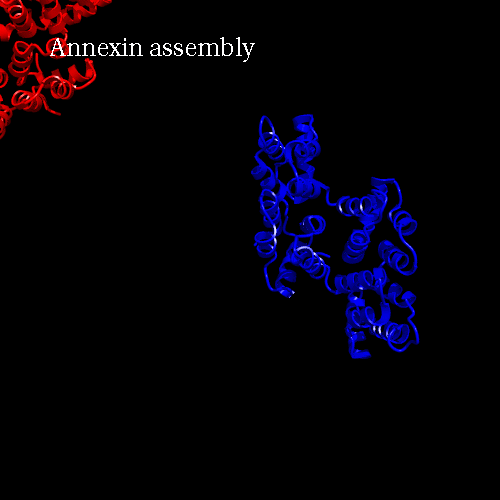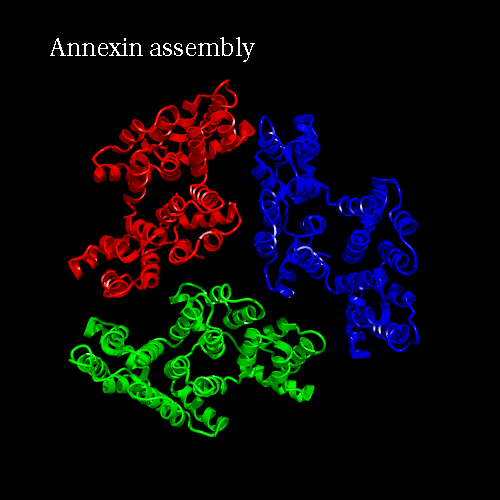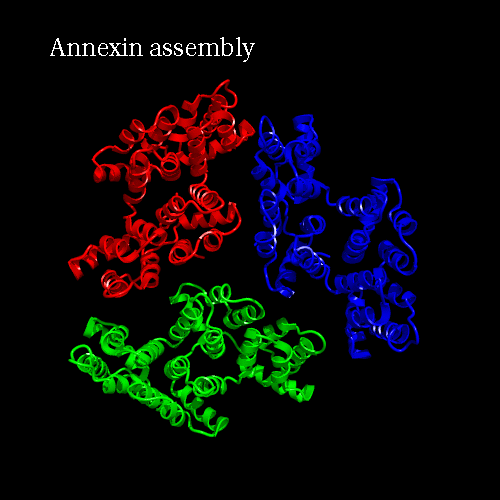
|
A more interesting example consists of using the "zoom" camera. In this case, we kept all the parameters of the previous representation, only changing the camera option to "zoom", the background to "t_spheresky2", and the positions of each chain (A, B, C) and the bounded form (A-B-C, "All") in the 3*3 panel as follow:
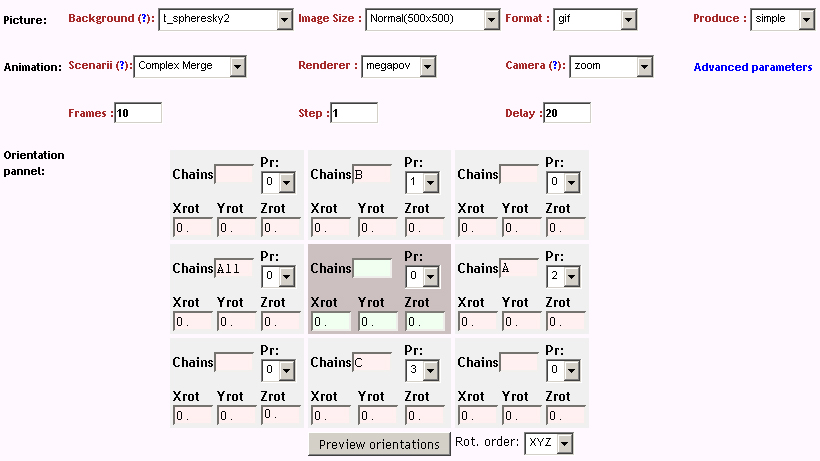
|
Engine : Dino / Render : Megapov
|
Engine : YASARA / Render : Megapov
|
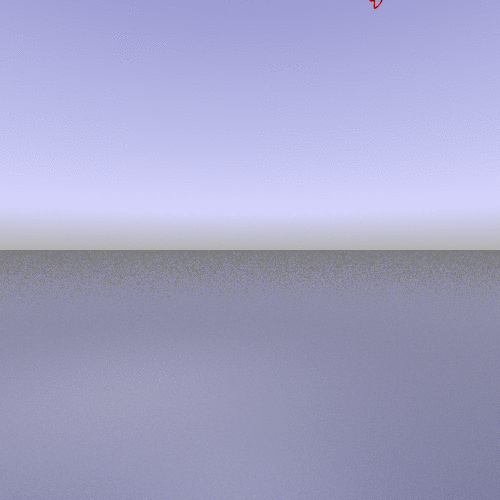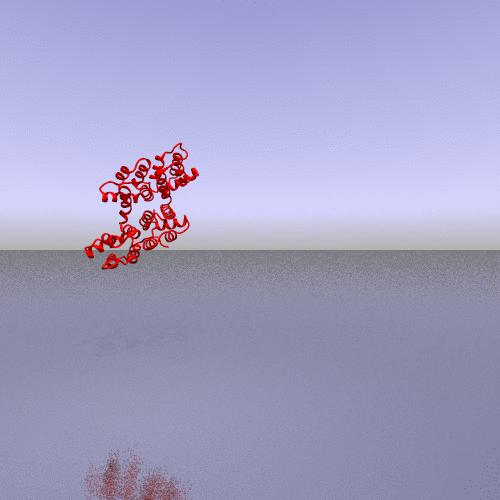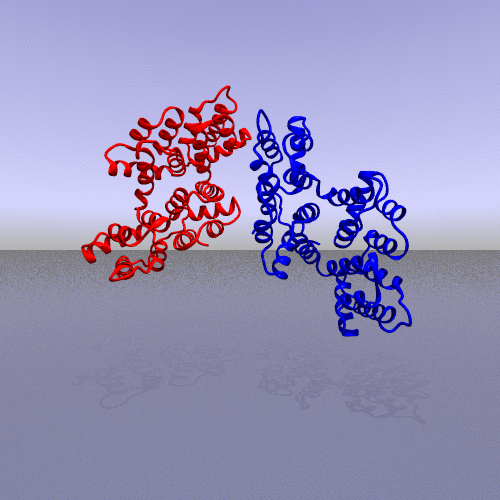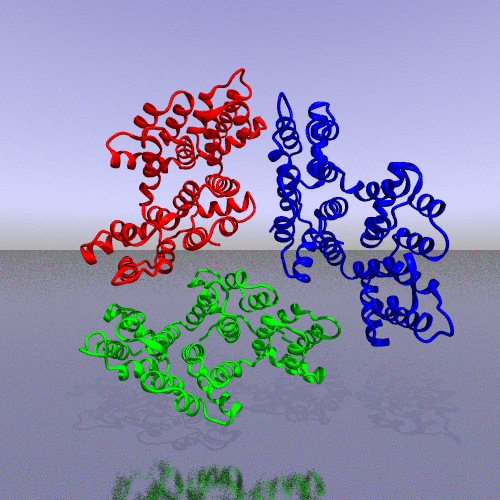
|
|
|