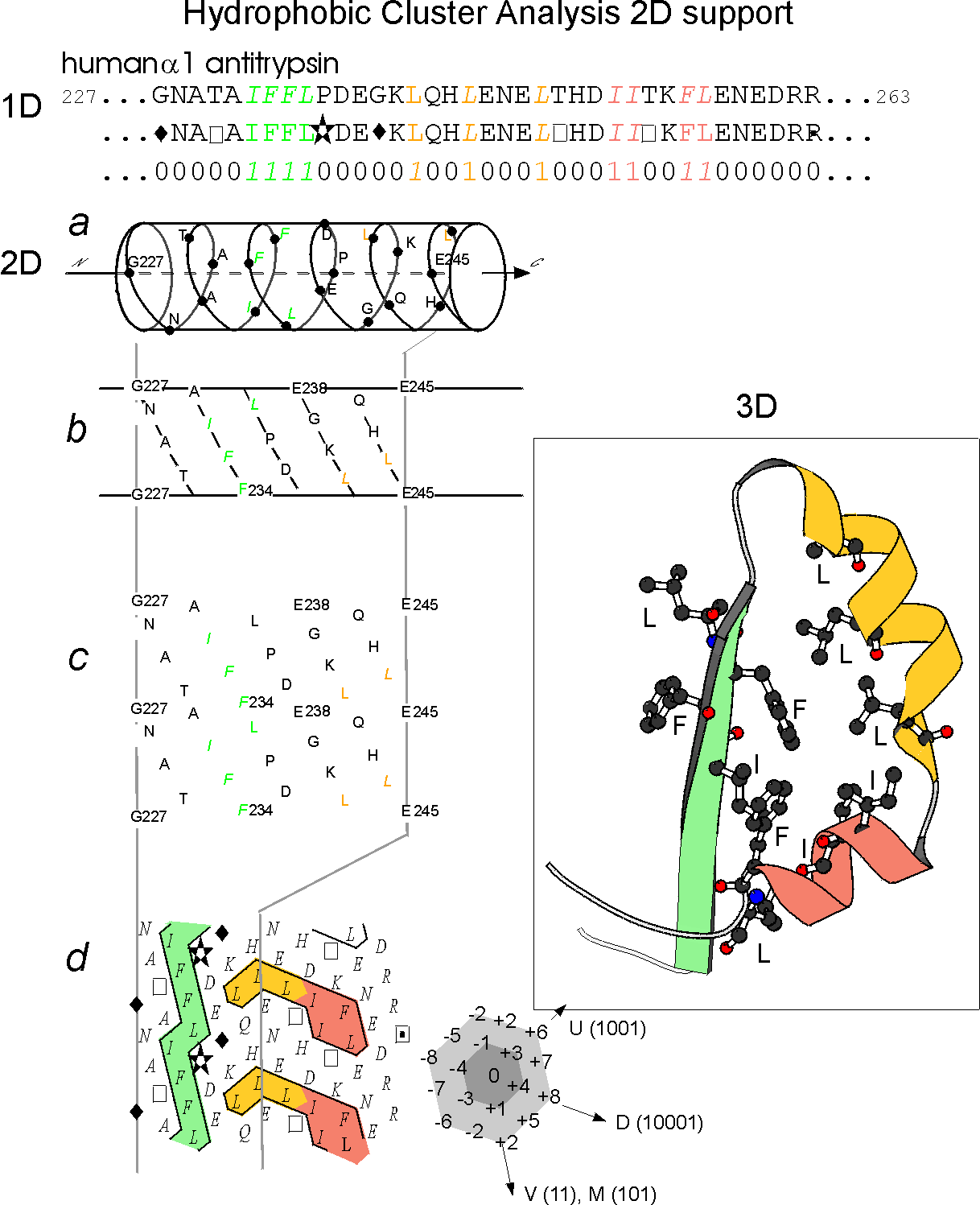
The HCA method is
based
on the use of a bidimensional plot, called the HCA plot and whose
principles
are illustrated on the following figure (Figure 1).
The bidimensional
plot is associated with an alpha helicoidal pitch (3.6 residue/turn,
connectivity
distance (residues separating two different clusters) of 4) which has
been
shown to offer the best correspondence between clusters and regular
secondary
structures (Refs. 1 and 2). Examination of the HCA plot of a protein
sequence
allow to easily identify globular regions from non globular ones and,
in
globular regions, to identify secondary structures. This 2D signature,
which is much more conserved than 1D sequence and which can be enriched
from the comparison of families of highly divergent sequences, allows
to
succesfully detect at low levels of sequence identity significant
similarities
(on the structural and functionals levels) from background noises.
An
example of the detection of a duplicated domain in a protein sequence
Two
examples of clusters often associated with beta-strands and
alpha-helices

More
details are available in :
1. Deciphering
protein
sequence information though hydrohobic cluster analysis. Current
status and
perspectives.
Callebaut I, Labesse G, Durand P, Poupon A, Canard L,
Chomilier J,
Henrissat
B., Mornon JP. Cell. Mol. Life Sci. (1997) 53,
621-645
2. Detection of
secondary
structure elements in proteins by hydrophobic cluster analysis.
Woodcock S, Mornon
JP, Henrissat B. Protein Eng (1992) 5 (7):
629-635
3. Hydrophobic
cluster
analysis: procedures to derive structural and functional
information from
2-D-representation
of protein sequences. Lemesle-Varloot L, Henrissat
B, Gaboriaud C,
Bissery
V, Morgat A, Mornon JP. Biochimie (1990) 72 (8):
555-574
4. Hydrophobic
cluster
analysis: an efficient new way to compare and analyse amino acid
sequences. Gaboriaud
C, Bissery V, Benchetrit T, Mornon JP. FEBS Lett
(1987) 224
(1):
149-155