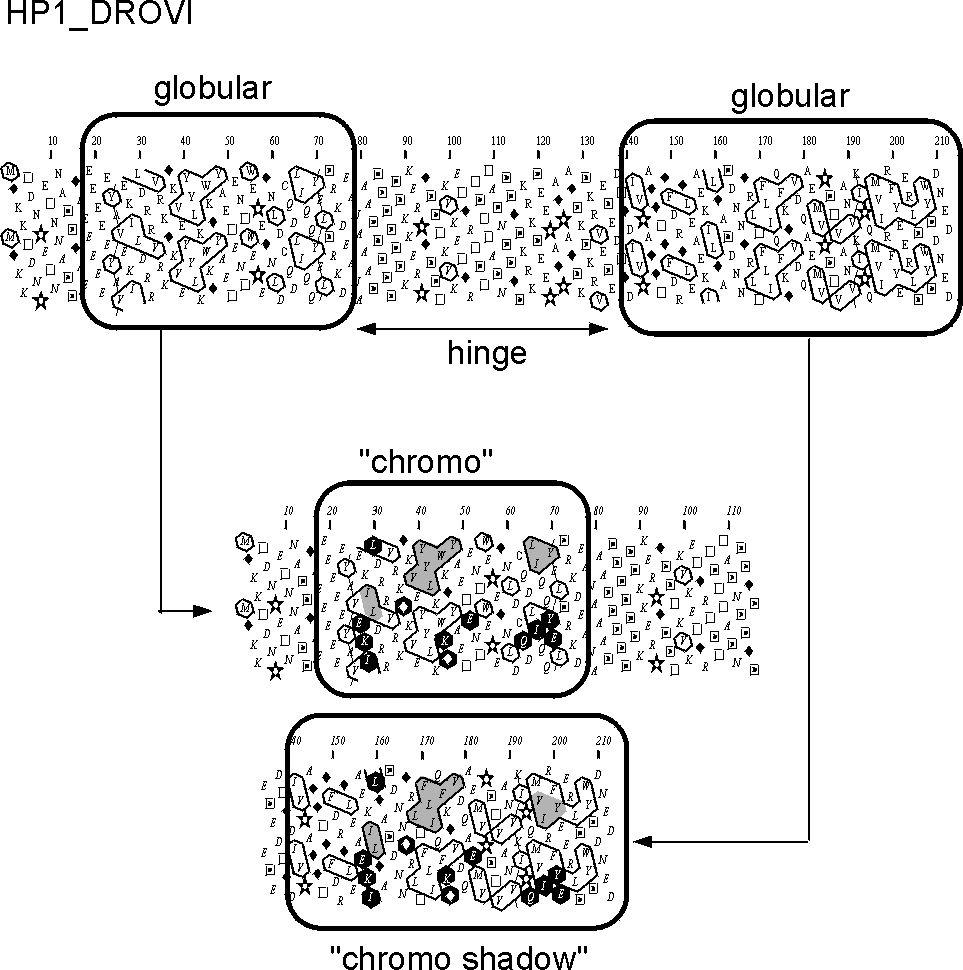

This example deals with the detection of a duplicated "chromo" domain in the sequence of the heterochromatin protein 1 HP1 (SwissProt identifier HP1_DROVI). Two globular domains can be easily identified (containing about 1/3 of hydrophobic amino acids), separated by a clear hydrophobic region (hinge). The two domains of similar length share similar clusters (shaded grey), associated with identities (~ 20 % identity). The hydrophobic core is clearly conserved between the two protein domains, as assessed by a high HCA score (% of hydrophobic amino acids which are topologically conserved (often not chemically identical)).
Details about chromo domains can be found in
1. Biochem
Biophys Res Commun 1997 Jun 9;235(1):103-7 Hydrophobic cluster
analysis reveals a third
chromodomain in the Tetrahymena Pdd1p protein of the
chromo superfamily. Callebaut I, Courvalin JC, Worman HJ, Mornon JP
2. J
Biol Chem 1997 Jun 6;272(23):14983-9 Domain-specific interactions of
human HP1-type
chromodomain proteins and inner nuclear membrane protein
LBR. Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ
as well as in :
(description of the chromo family)
3. Nucleic
Acids Res 1995 Aug 25;23(16):3168-74 The chromo shadow domain,
a second chromo domain
in heterochromatin-binding protein
1, HP1. Aasland R, Stewart AF
4. Nucleic
Acids Res 1995 Nov 11;23(21):4229-33 The chromo superfamily:
new members, duplication of
the chromo domain and possible
role in delivering transcription regulators to chromatin. Koonin EV, Zhou
S, Lucchesi JC
(review)
5. Curr
Opin Cell Biol 1998 Jun;10(3):354-60 Chromo-domain proteins:
linking chromatin structure
to epigenetic regulation. Cavalli G, Paro R (review)
(3D structure)
6. EMBO
J 1997 May 1;16(9):2473-81 Structure of the chromatin binding (chromo)
domain
from mouse modifier protein 1. Ball LJ, Murzina NV, Broadhurst
RW, Raine AR, Archer SJ, Stott FJ, Murzin AG,
Singh PB, Domaille PJ, Laue ED