

Next: SCatmMiss ?
Up: Methods of the PDB
Previous: CA only ?
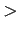 x.BBatmMiss()
x.BBatmMiss()
This will return the number of amino-acid residues having some backbone
missing atom (one of N, CA, C, O), and a string concatening the information
about all
the residues with missing peptidic chain atoms. For each residue,
the string consists of RName_ChLbl_RNum_RIcode, where RName is the name of the
residue (3 letters), ChLbl is the chain label, RNum is the number of the residue (as in the PDB file) and RIcode the PDB insertion code of the residue.
Apache
2004-03-20