

Next: advanced amino acid sequence
Up: Advanced features related to
Previous: advanced chain information
Fragments are defined as series of amino-acids that are covalently bonded, and are not
bonded to other fragments.
A series of methods are related to their detection. The generic method is frgList.
Usage:
frgList(maxNCDist = 1.7, maxCADist = 4.1)
This will return the number of fragments detected, and their boundaries (numbered from 0).
This method will detect fragments onthe basis of the inter-atomic distance between the C' atoms of residue 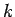 and the N atom of residue
and the N atom of residue 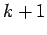 . If only the CA atoms are provided,
the distance between the consecutive alpha-carbons will be used instead. If not specified,
the default inter-atomic distance provided are of 1.7 and 4.1. The values were chosen since found to encompass much of the PDB deviations to standard geometry. They can be modified as
any named parameter.
. If only the CA atoms are provided,
the distance between the consecutive alpha-carbons will be used instead. If not specified,
the default inter-atomic distance provided are of 1.7 and 4.1. The values were chosen since found to encompass much of the PDB deviations to standard geometry. They can be modified as
any named parameter.
The more advanced methods wrapped by frgList are: chnCAFrgList() (for CA fragments) and
chnFrgList() (for all atom fragments).
The nFrg() method will only return the number of fragments.


Next: advanced amino acid sequence
Up: Advanced features related to
Previous: advanced chain information
Apache
2004-03-20