query: 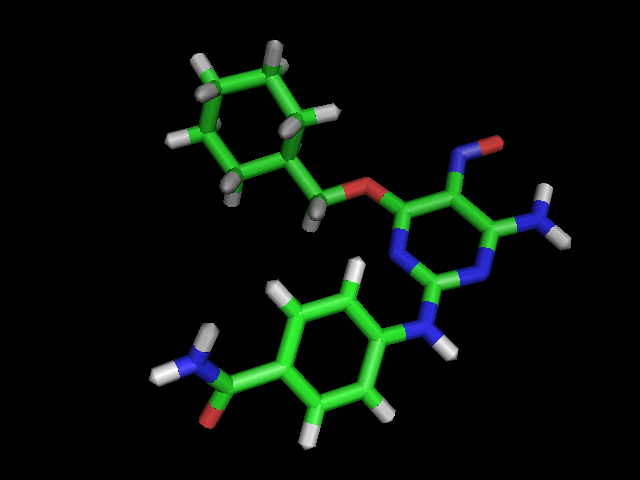 |
==> | similar
compounds: 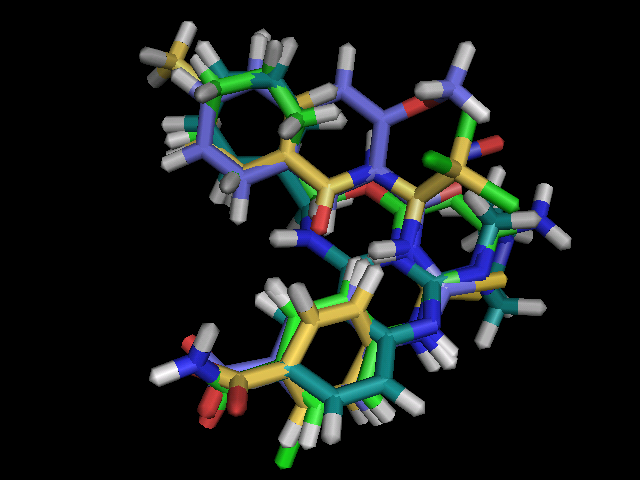 |
Access the service
Note on navigator: a strange behaviour of the service has been noted on Safari (at least on Leopard MacOS X). Results sometimes do not appear properly. Please prefer Firefox.
Note on jmol applet: on the result page, the jmol applet will sometimes not load properly, depending on the browser. We suspect this might come from interferences between ajax and jmol. Usually, the direct access to the result page noted in green on top (e.g. http://mobyle.rpbs.univ-paris-diderot.fr/tmp/wwLig-CSRre/U30250938369036) will solve this.
Note on execution times: wwLigCSRre typical execution time may vary depending on server load and bank size, from few minutes to several hours.
Yet, the server offers the possibility to send an email to notify job completion. Not using this facility, you may also click on the "update job status" to refresh the page
(see the video tutorial available on top of the Mobyle portal).
1. History
2. Features
3. Limitations
4. Usage
5. Some tools of get mol2 files
6. Examples, sample tests
7. Concepts
8. Validation
9. Availability
History:
- 2006: early discussions at RPBS about coupling the CSR algorithm with some regular expression patterns (REs) to prune the CSR 3D similarity search algorithm.
- 2006: First implementation of the CSRre algorithm in the scope of proteins.
- mid-2008: Implementation of LigCSRre: derivation of REs for small
compounds, based on the mol2 atomic types.
- August 2008: wwLigCSRre server opened for internal use at RPBS. Intensive tests on compound enrichment on the Chembridge diversity set. LigCSRre proves efficient for compound enrichment.
- October 2008: Service opened to the
community for small banks only, due to calculation cost.
- February 2009, 9th: small bug fix for very small bank size (e.g. 1 conformation only).
- Given a query (mol2 format), the server will scan a bank of
compounds (user selected) and return compounds ranked by similarity.
- The server will also return the top compounds superposed onto the
query, for further investigation by users.
Limitations:
- LigCSRre is more time consuming than 2D similarity methods for
instance. Hence, for wwLigCSRre we limit bank size in terms of
compounds. Bank
compounds are multiconformers (up to 50 per compound). More banks to be
available.
- Physico-chemical rules used for this server have proven
efficient, but could be optimized to introduce more biological/chemical
knowledge for compounds (e.g. atoms of particular importance).
- The LigCSRre scores have no statistical significance (as most
methods of the field), other than a zScore based on the numberof bonds
paired. A visual
inspection of the results by a medicinal chemist is highly recommended.
Usage:
- The demonstration mode will launch the program with the test
data described below (Examples). The content of input data fields
will not be considered.
- Input: one compound (mono
conformation) in the mol2 format.
- Processing: the user can choose among several banks (last update
January 2009).
- "small test set": This is a set of 47 active compounds on Thyminide Kinase, RNAse, CDK2, FXa, and NA, left for tests (used for demonstration).
- Aurora Fine Chemicals focused collections:
- Analgesic compounds : This is a subset of 1587 compounds of
the collection, using a Tanimoto diversity criterion of 0.8.
- Antibacterial compounds: The complet antibacterial
collection, including 2069 compounds
- Anticancer compounds: This is a subset of 2048 compounds of the collection, using a Tanimoto diversity criterion of 0.8.
- Ion-channel: This is a subset of 775 compounds of the collection, using a Tanimoto diversity criterion of 0.8.
- Kinases: This is a subset of 2283 compounds of the collection, using a Tanimoto diversity criterion of 0.8.
- Chembride collections:
- Diversity set: This is a 2880 compound subset of the
Chembridge diversity set (39623) molecules, filterered using cactus
using a diversity index of 0.76.
- CNS (Central Nervous System): This is a subset of 2363 compounds of the collection, using a Tanimoto diversity criterion of 0.75.
- G Protein Coupled Receptors (GPCR): This is a subset of 940 compounds of the collection, using a Tanimoto diversity criterion of 0.85.
- DrugBank collections:
- Small Molecules: This is a subset of 942 compounds the
DrugBank set
of 4857 compounds, obtained by ADME/tox filtering by FAFDrugs2 (soft
filtering).
- Approuved: this subset of 409 compounds resulting from ADME/tox filtering by FAFDrugs2 (soft filtering) have been approved by the FDA.
- Withdrawn: these 24 compounds compliant with ADME/tox filtering by FAFDrugs2 (soft filtering) have been withdrawn from public availability due to some side effects.
- DSSTOX:
- Carcinogenic potency: This set is derived from the Environmental Protection Agency CPDBAS (Carcinogenic Potency Database) set. This set includes 1551 potentially toxic compounds.
- It is possible to upload a bank, but a maximum of either 500
compounds
or 10,000 conformers wichever is limiting, is accepted.
- The multiconformations are detected on the basis of the _1, _2, etc added to the compound name (e.g. the compound of id 11233452 has conformer ids on the form 11233452_1, 11233452_2, ...)
- Using this upload possibility, wwLigCSRre can also be used just to superimpose small compounds, submitting all but one compound as the bank, and the remaining one as query.
- The search is tuned using hidden parameters values validated on the complete Chembridge diversity set.
- It is possible to disable the use of internal equivalence classes (see the concepts section) that define generic atomic type equivalences. Swithcing off this option will revert to accept pairing of atoms having the exact same mol2 type.
- It is possible to tune the number of 3D superimpositions returned
in the PDB format.
- Output: The server will return a score file ( a file of bank compounds ranked by similarity scores), and a mol2 file containing the best compounds superposed onto the query.
- the score file will report information per compound of the bank, lines are sorted so that first lines correspond to best matches, and last to worse:
- Compound Id, number of bonds paired, (query / compound number
of bonds), number of atoms paired, RMSd over the paired atoms, zScore
based on the number of bonds shared.
- zScore values more than 2 are significant (but some active
compounds have zScore values less than 2).
- It is important to compare the number of paired bonds to the
theoretical maximum (all bonds match) on both the query and the match
to get an idea of the similarity.
- It is also important to
visualize the superimpositions of the compounds.
- the mol2 file contains the bank compounds superimposed
onto the query, according to the paired atoms.
The mol2 format should be on the form below (More explanations on the format here). A tool to convert from most formats to mol2 is here.
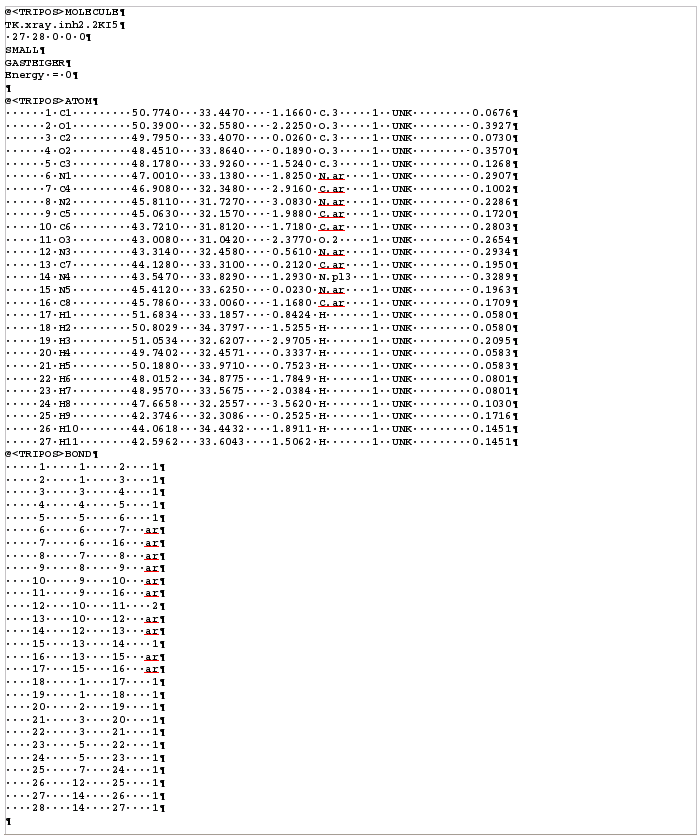
Some tools to get mol2 files:
- openbabel is a program to convert from other formats to mol2.
- JME is tool to draw a compound and get its smiles representation.
- Frog is a tool to generate 3D from smiles.
- External sites such as Zinc and PubChem also propose facilities to retrieve compounds.
1. CDK2 inhibitor
A query compound (CDK2 inhibitor) to test the server is accessible here.
Run against the "small test set", the resulting score file is here, and the resulting PDB file is here.
Note: From one run to another, the results may slightly differ since LigCSRre is a stochastic approach.
The 3D superposition obtained for CDK2 compounds:
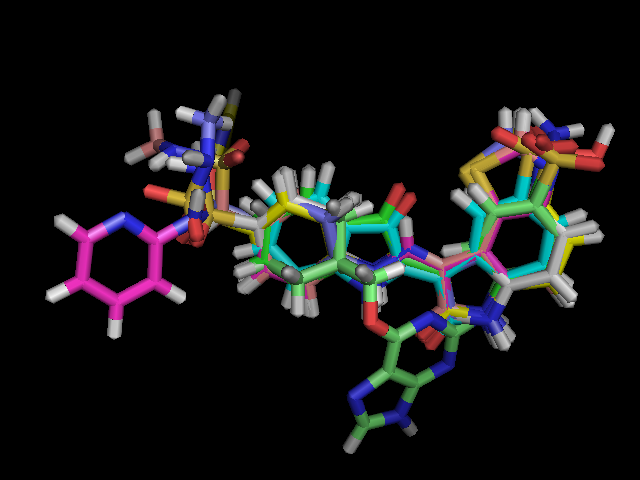
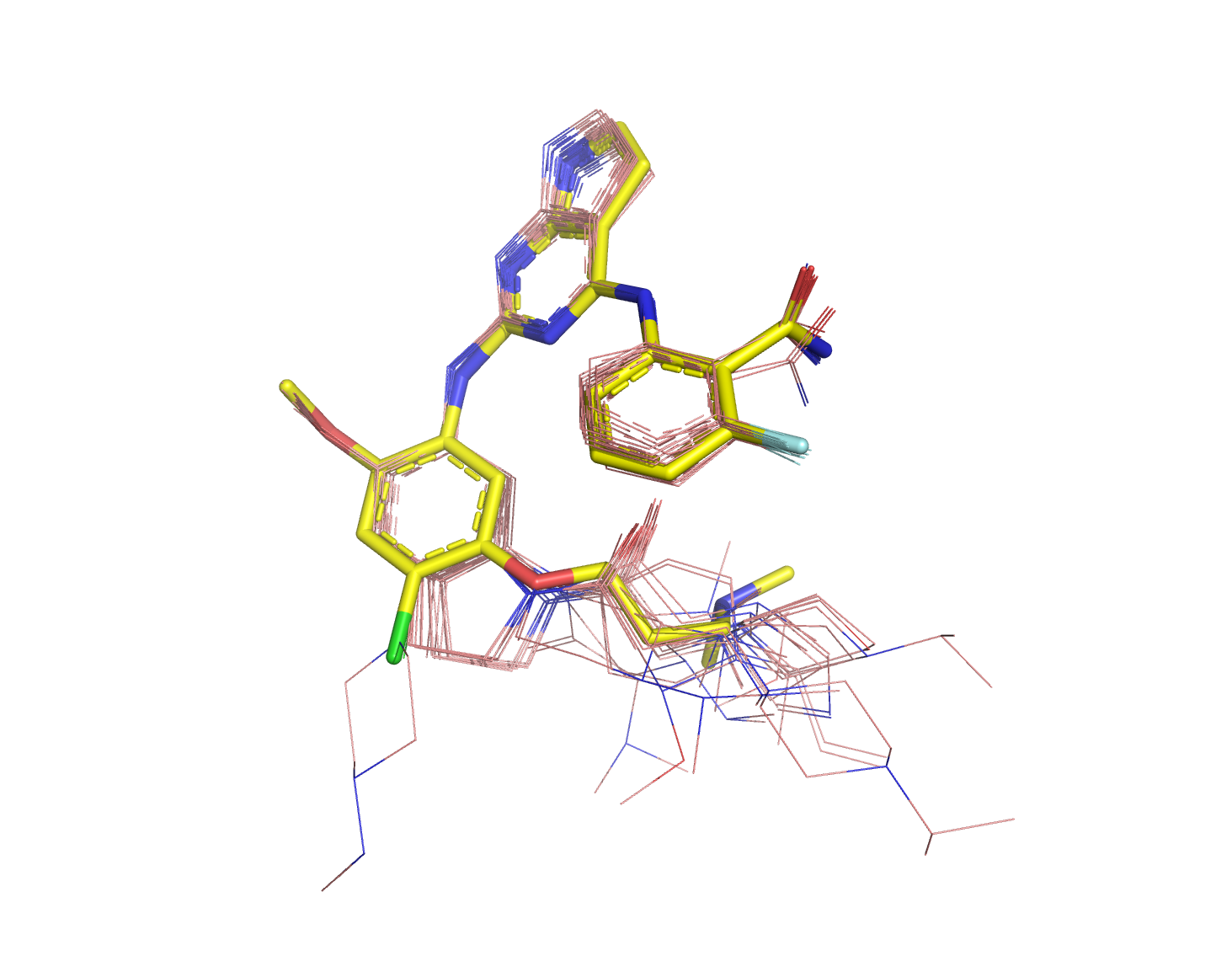
2. Structure Activity Relationship study on Insulin like Growth Factor - 1 Receptor
A query compound to run against this SAR-IGF-1R set is accessible here.
Run against the "small test set", the resulting score file is here, and the resulting mol2 file is here.
Note: From one run to another, the results may slightly differ since LigCSRre is a stochastic approach.
The 3D superposition obtained:
- CSR algorithm. More details are available from the publication of the original CSR algorithm.
- Regular expressions: LigCSRre relies on a three stage regular expression mechanism.
- 1. Default rules : pairings are allowed only with atoms having the exact same mol2 atomic type.
- 2. Generic equivalence classes: pairings are allowed between atoms of the same equivalence class.
- LigCSRre internal equivalent classes are:
- carbons but carbo-cations
- sp2 Oxygens (0.2 and 0.co2 mol2 types)
- Sulfoxide and sulfone Sulfurs (S.o and S.o2)
- sp2 and sp3 Sulfurs
- Nitrogens
- 3. File specific rules (Not yet available from the server).
- Precedence is set as Default rules < Equivalent classes < File specific rules
- The file defining the equivalence classes is here.
CSR: M.
Petitjean "Interactive Maximal
Common 3D
Substructure Searching with the Combined SDM/RMS Algorithm"
Comput. Chem. (1998) 22[6],463-465).
Validation tests:
- Validation tests have been performed on the complete Chembridge diversity set (50000 compounds), filtered for ADME/Tox compliance. This resulted on a subset of 37907 compounds.
- Tests have been made for all the 47 compounds of the small test set of this service.
- Enrichment results show, over these ligands, that LigCSRre
performs slightly better than related softwares for early enrichment
(Quintus, Sperandio et al., submitted).
Availability:
- It is possible to contact the authors to run the software on
larger banks (e.g. complete diversity set), on specialized banks (e.g.
kinase, GPCR, ...), or on private banks.