PatchSearch
A service to identify structurally conserved regions at the protein surfaces.
A service to identify structurally conserved regions at the protein surfaces.
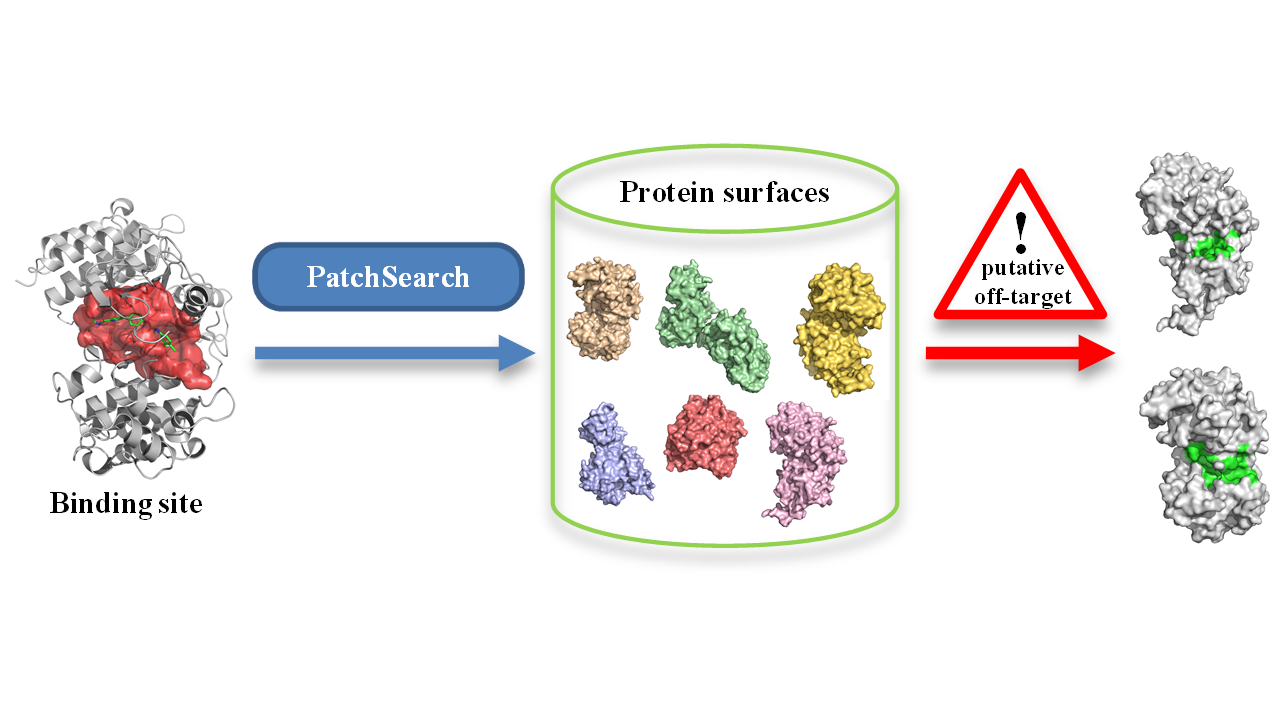
Many therapeutic molecules are known to bind several proteins, which can be different from the initially targeted one. Such unexpected interactions with proteins called off-targets can lead to adverse effects. Potential off-target identification is important to predict to avoid drug side effects or to discover new targets for existing drugs.
This service implements the PatchSearch method [1], which allows local nonsequential searching for similar regions, called patches, on protein surfaces. It is based on detection of quasi-cliques in product graphs representing all the possible matchings between a patch and compared structures.
The service is fully embedded in the Mobyle framework. It embeds simple yet powerful data management features that allow the user to reproduce analyses. It gives to the users the possibility to create registered accounts, which allows user data and jobs to be maintained and managed across multiple work sessions and therefore to reuse and share data and results.
Access the service through the RPBS Mobyle portal:
When using this service, please cite the following references:
PatchSearch: a web server for off-target protein identification.
Nucleic Acids Res. 2019 Jul 2;47(W1):W365-W372.
PatchSearch: A Fast Computational Method for Off-Target Detection.
J Chem Inf Model. 2017 Apr 24;57(4):769-777.
The Web server requires three inputs:

This section will incrementally provide information about job progression and errors if any. A typical run should produce a report similar to the following :

Errors related to the input data specified are also reported in this field.

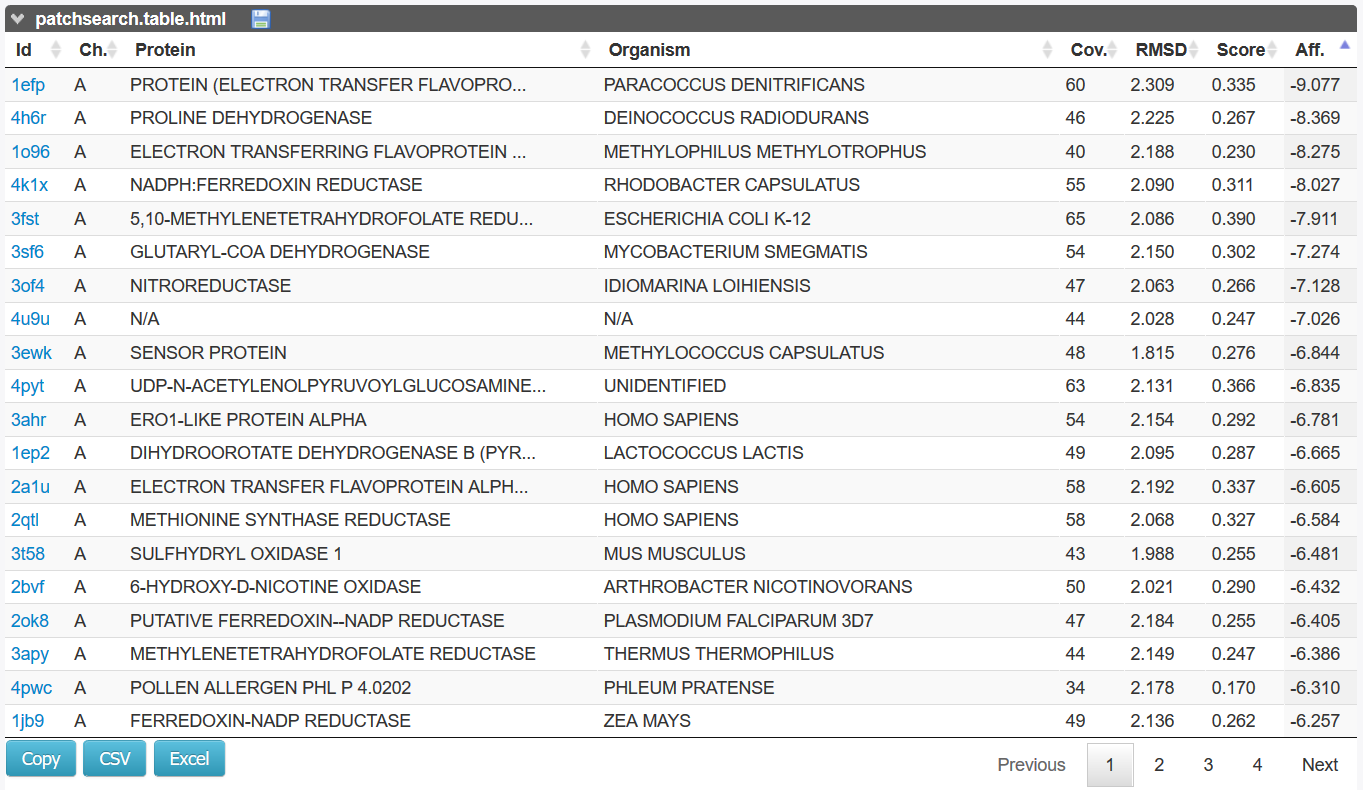
PatchSearch results is a table for each retrieved protein.
Similar patches are output to a table with the PDB identifier, the coverage (number of retrieved atoms), the RMSD (between the query patch and the retrieved patch), the PatchSearch Score (relative Binet-Cauchy score) and the affinity computed by a scoring step using Smina program [2].
The relative Binet-Cauchy score is based on the BC-score [3] weighted by the proportion of the number of retrieved atoms to the number of query atoms).

An interactive page allowing to browse the retrieved patches. The residues forming the patch are in thin stick representation with the carbon atom in green. The ligand is in thick stick representation with the carbon atom in cyan. The user can visualize the each solutions ranking to the estimated affinity by Smina program using Vinardo scoring function [4].
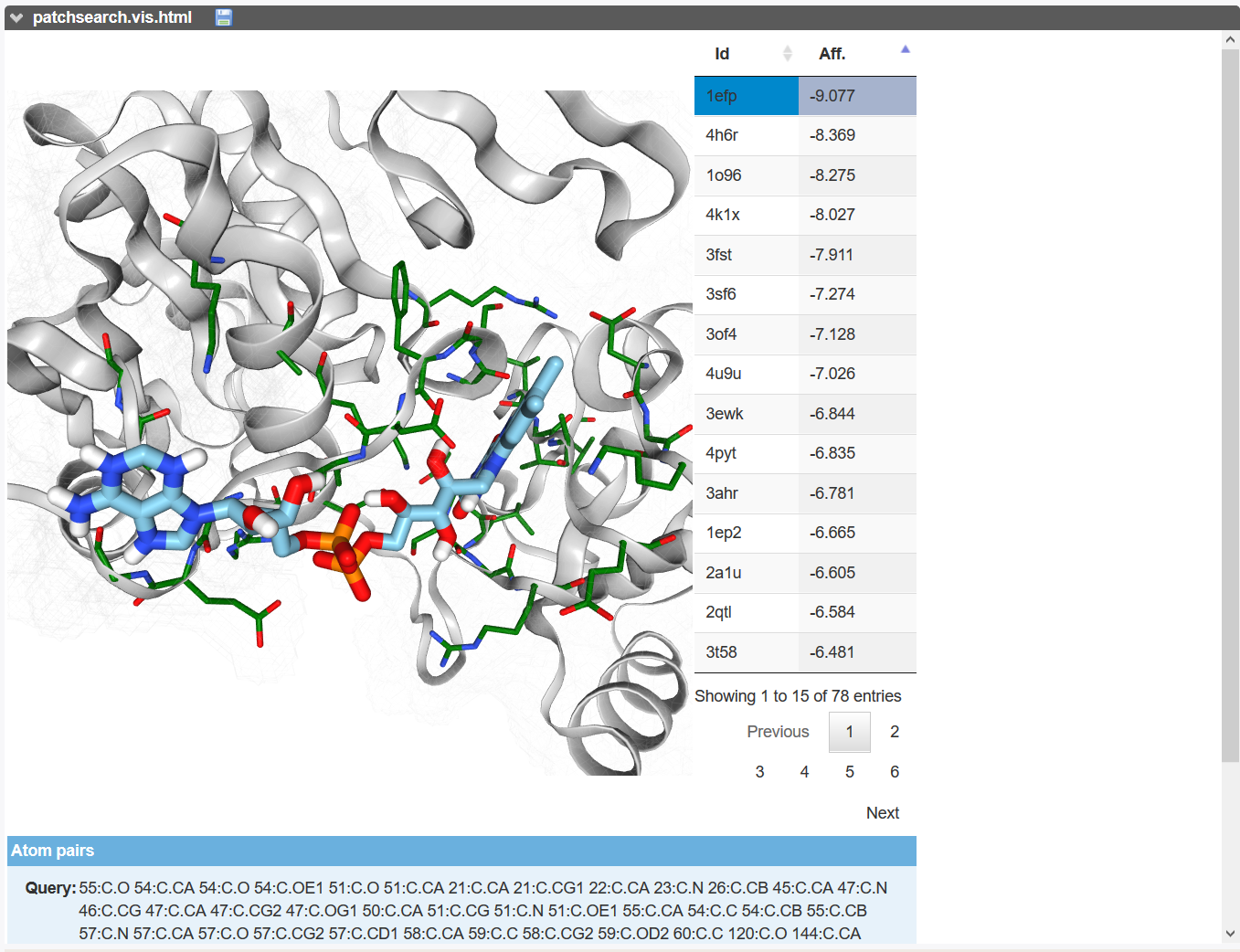

The table of results is downloadable in csv or Excel format.
In this example, we aim to detect proteins which may interact with Naproxen. We used the structure of bovine lactoferrin complexed with Naproxen as query patch against a list of 20 proteins structures.
As a simple test, you can:
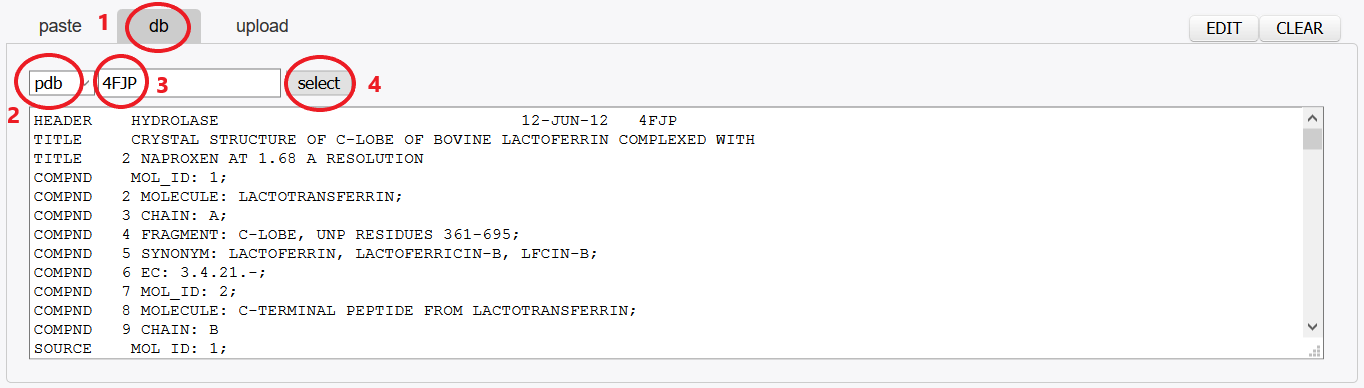
Use Mobyle facilities: fetch a protein structure from the PDB databank using a PDB identifier.
Click the db button.
Select pdb in the menu.
Enter a PDB identifier: 4FJP.
Press select button.
Define the patch:
Press Ligand visualization button.
Select the ligand identifier, [NPS]711:A, in the list on the right side of the graphic window.
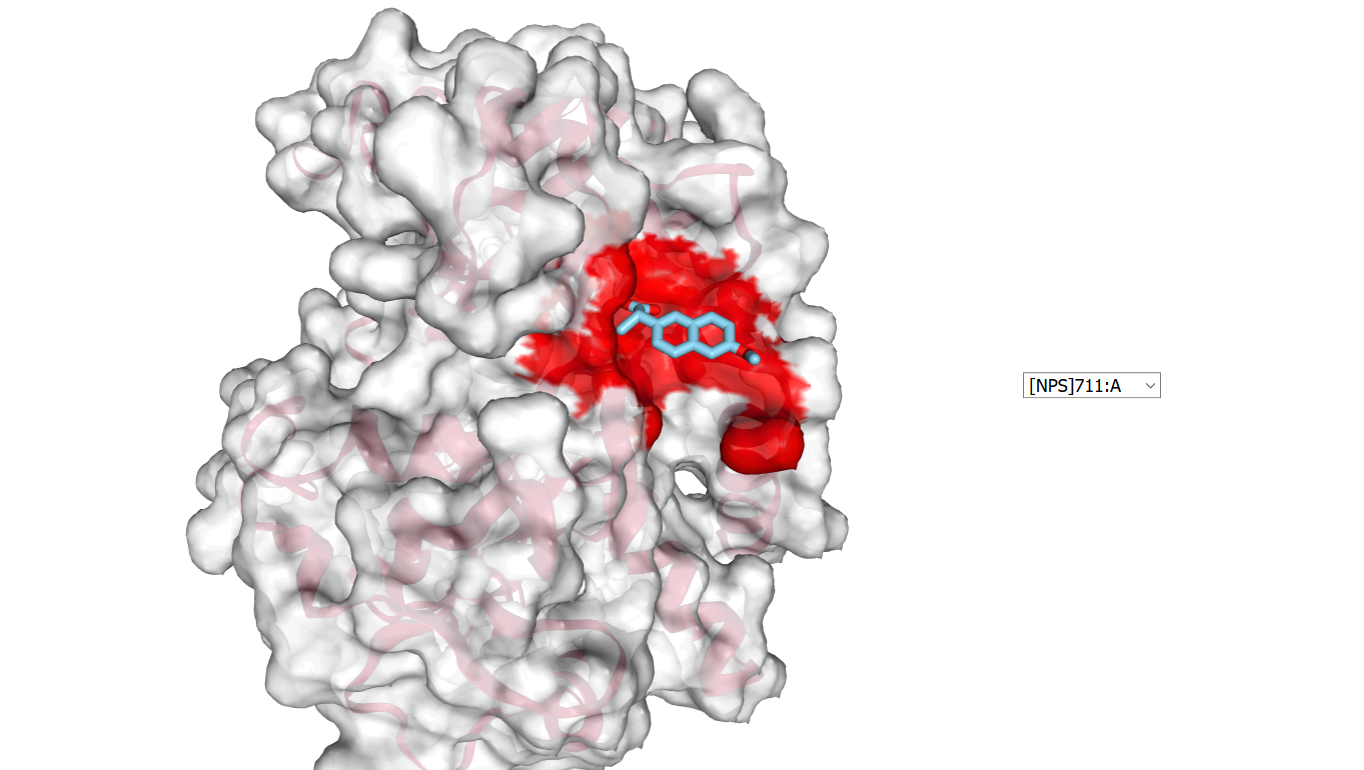
Define the targeted proteins:
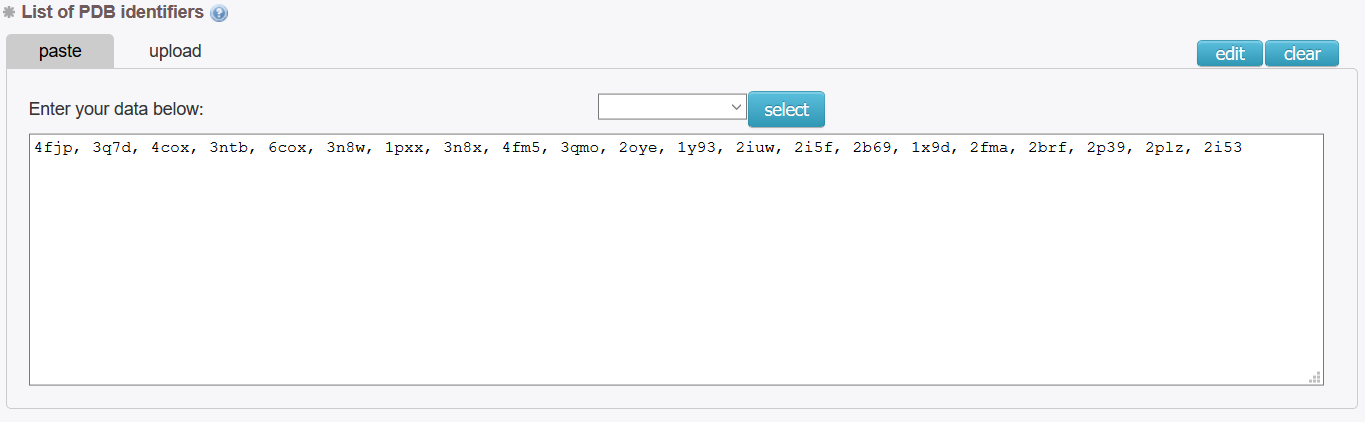
Input a list of targeted proteins into the field:
4fjp, 3q7d, 4cox, 3ntb, 6cox, 3n8w, 1pxx, 3n8x, 4fm5, 3qmo, 2oye, 1y93, 2iuw, 2i5f, 2b69, 1x9d, 2fma, 2brf, 2p39, 2plz, 2i53
Run:
Launch PatchSearch by clicking Run either at the top or at the bottom of the page.
Results:
The results of the search can be accessed here.
[1] Rasolohery I, Moroy G, Guyon F.
PatchSearch: A Fast Computational Method for Off-Target Detection.
J Chem Inf Model. 2017 Apr 24;57(4):769-777.
[2] Koes DR, Baumgartner MP, Camacho CJ.
Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise.
J Chem Inf Model. 2013 Aug 15;53(8):1893-904.
[3] Guyon F, Tufféry P.
Fast protein fragment similarity scoring using a Binet-Cauchy kernel.
Bioinformatics. 2014 Mar 15;30(6):784-91.
[4] Quiroga R, Villarreal MA.
Vinardo: A Scoring Function Based on Autodock Vina Improves Scoring, Docking, and Virtual Screening.
PLoS One. 2016 May 11(5):e0155183.