Welcome to the GLSearch structural similarity search server!
GLSearch is a fast and flexible approach aimed at identifying in large collections of structures linear fragments similar to a query. GLSearch is based on a new similarity kernel approach, scrogin similarity between -1 and 1, where a value of 1 corresponds to the exact same conformation than the input, and -1 to the mirror conformation.
 |
 |
 |
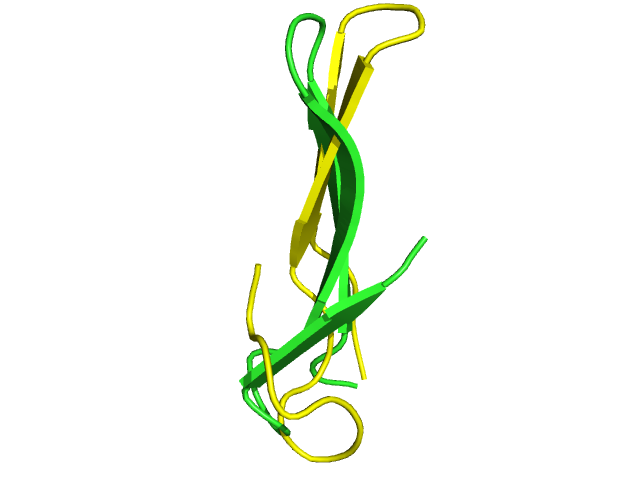 |
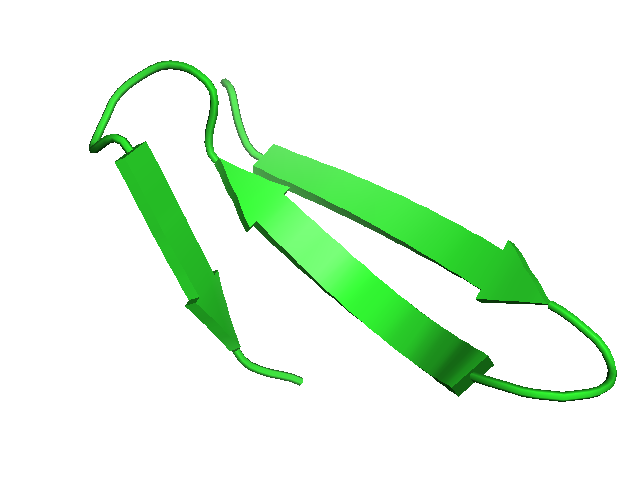
| 2zaj query fragment. | 24 matches identified in Astral-1.75 at a score better than 0.75, and accepted deformation of 0.3. RMSd vary between 1.9 and 4.3 Angstroms. | Matches identified using RMSd alone as a filter, for a maximum RMSd of 4 Angstroms. | d1vpra1 fragment having a mirror conformation to that of 2zaj (beta-sheet twist is inverted. |
Access the
GLSearch service @ the
RPBS Mobyle Portal.
When using this service, please cite the following references:
A kernel approach to fast protein fragment mining.
submitted
Minimal fragment size if 5 residues.
When using this service, please cite the following references:
A kernel approach to fast protein fragment mining.
submitted
Features
- Ungapped fragment similarity search: Starting from a query, mine large collections of structures to identify similar fragments. On request, get both fragments and sequences in return.
- Search constraints: GLSearch makes use of 2 parameters to drive the search. (i) The kernel score, betwen 1 (perfect match) and -1 (perfect mirror). (ii) The fragment deformation: since the kernel score if flexible, a maximum deformation makes easy to narrow the matches in the vicinity of the query.
- Recursive similarity: Finely tune the shape of the matches using the kernnel score at a local level (subfragments) in addition to the complete fragment level. This ensures each local part of the matches are similar to the query. Note: this can be misleading.
- Mirror search facility: In addition to searching for similar fragments, you can as well search for mirror conformations.
Limitations
- Fragment size: Presently, the search is based on alpha carbon coordinates. Too small fragments (less than 5 amino acids) can lead to unreliable results. There is in theory no upper limit, but the probability to identified long similar ungapped ffragments is low ...
Usage
Input
- Pre-configured test: By setting this option to "Yes", GLSearch will be run using fragment 16-40 of PDB 2zaj entry (a WW domain), searching for matches with a kernel score more than 0.75 and a fragment deformation not exceeding 30%.
- Input structure: Input query file must be in PDB format.
- Searching a subpart of the query: It is possible to restrain the serach to a subpart of the input PDB, by specifying the numberof the first and last residues (numbered from 1) delimiting the subpart of the structure.
- Bank to mine: Banks to mine correspond to subsets at 70% sequence identity of Astral 1.75 [1] and the culled PDB [2].
Search parameters
- Pruning the search: Matches are identified depending on a combination of several conditions. The minimal global score specifies the minimal value of the kernel sccore of the match. Proposed values are between 0.2 and 1, where 1 is the perfect structural identity. The maximal deformation rate specifies the upper limit of the flexibility, between 0. and 1., where 1. means no constraint and 0 stands for no deformation. It is in addition possible to specify a minimal local score, to recursively constrin the similarity at the local level of the query. It is achieved using sliding window of seven residue length. Matches for which at least one of the local sccore is less than the minimal value will be discarded. Note this can be very misleading, particularly for beta stranded fragments.
- Mirror conformations: Several level of anti-symmetry can be considered. Setting both direct and mirror, matches with scores either more than the minimal global score OR less than minus the minimal global score will be returned. Setting mirror only, only the matches with scores less than minus the minimal global score will be returned. At the local level, local mirror conformations corresponds to both direct and mirror matches.
- Output options: Switching Superimpose to Yes, GLSearch will extract each fragment and superimpose it onto the query. Since this can be very long compared to the search, (i) it is not the default choice, and (ii) only the Max. matches best matches (sorted by decreasing kernel score) will be returned.
Results
-
Progress report
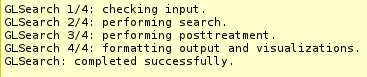 TGLSEarch incrementally returns information about job
progression and errors if any, although typical run times are on the order of few seconds.
A typical run should produce a report similar to that. Errors related
to the input data specified are now also reported in this section.
TGLSEarch incrementally returns information about job
progression and errors if any, although typical run times are on the order of few seconds.
A typical run should produce a report similar to that. Errors related
to the input data specified are now also reported in this section.
-
Visualization of best matches
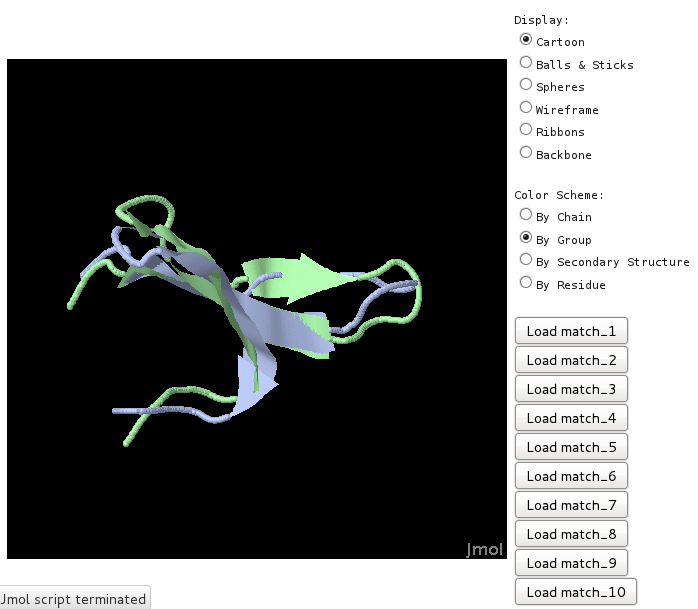 Up to the 10 best matches can be visualized superimposed onto the query using the Jmol applet [3]. This is only efffective settting the Superimpose swith to Yes.
Up to the 10 best matches can be visualized superimposed onto the query using the Jmol applet [3]. This is only efffective settting the Superimpose swith to Yes.
-
Logo representation of the match sequencesA lgog representation of the squences of the matches is provided in order to provide some insight about sequence variability of the matches. This is only efffective settting the Superimpose swith to Yes.

-
Matches
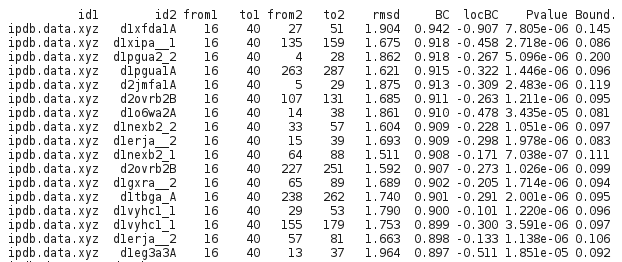 The matches are returned sorted by decreasing values of the kernel score.
For each match, the query and match PDB entry are reported together with their limits (from1, to1, from2, to2), their RMSd (as a supplementary information), their kernel score (BC), their worse local score (locBC), their p-value, and their deformation rate (Bound.).
The matches are returned sorted by decreasing values of the kernel score.
For each match, the query and match PDB entry are reported together with their limits (from1, to1, from2, to2), their RMSd (as a supplementary information), their kernel score (BC), their worse local score (locBC), their p-value, and their deformation rate (Bound.).
-
Match archive
 This archive contains the PDB fragments of the matches identified, if any, and for the Superimpose switched to Yes. It is in the unix tar format compressed using gzip.
This archive contains the PDB fragments of the matches identified, if any, and for the Superimpose switched to Yes. It is in the unix tar format compressed using gzip.
Once saved on your computer, enter for instance (unix) tar xzf GLSearch-matches.tgz to inflate the archive.
History
-
2010, September 10th - Initial development of the kernel score.
- 2011, nov 17 - First draft of a global approach to mine large collections fo structures.
- 2012, sep 1 - First implementation of the GLSearch service (restricted access). Internal tests.
- 2013, Jan 3 - First release of the service, open to all.
Known problems and answers
There is presently very few feedback reporting problems with GLSearch.
Most are related to Java and browser dependent behavior.GLSearch has been tested successfully using various OS / browser combinations, including:
Firefox 11 and Chrome 18 under Linux,
Internet Explorer 9, Firefox 3.6 and 11, Chrome 18 under Windows 7, Firefox 7 and Internet Explorer 8 under Windows XP,
Safari 5.1.(2,3), Firefox 11 and Chrome 18 under MacOS X (lion, snow leopard).
-
Jmol and OpenAstex applet/viewer will not launch properly In most cases, it is related to (i) security issue related to Java : check for messages asking for permission to launch the applet (ii) Java virtual machine does not behave properly: presently, please prefer official sun/oracle JRE or JDK to other implementations such as openJDK.
-
Results will not display on GLSearch termination - Mobyle upload button will not work. This has been observed depending on versions of the browsers, and seems to have been solved. If you encounter such behavior, just perform a full refresh of the result page (press Ctrl R or press Enter in the url specification field of the browser).
References
[3]
Jmol - a paradigm shift in crystallographic visualization.
Journal of Applied Crystallography, 43(5): 1250-1260 (2010).
<Back to top>
Jmol - a paradigm shift in crystallographic visualization.
Journal of Applied Crystallography, 43(5): 1250-1260 (2010).